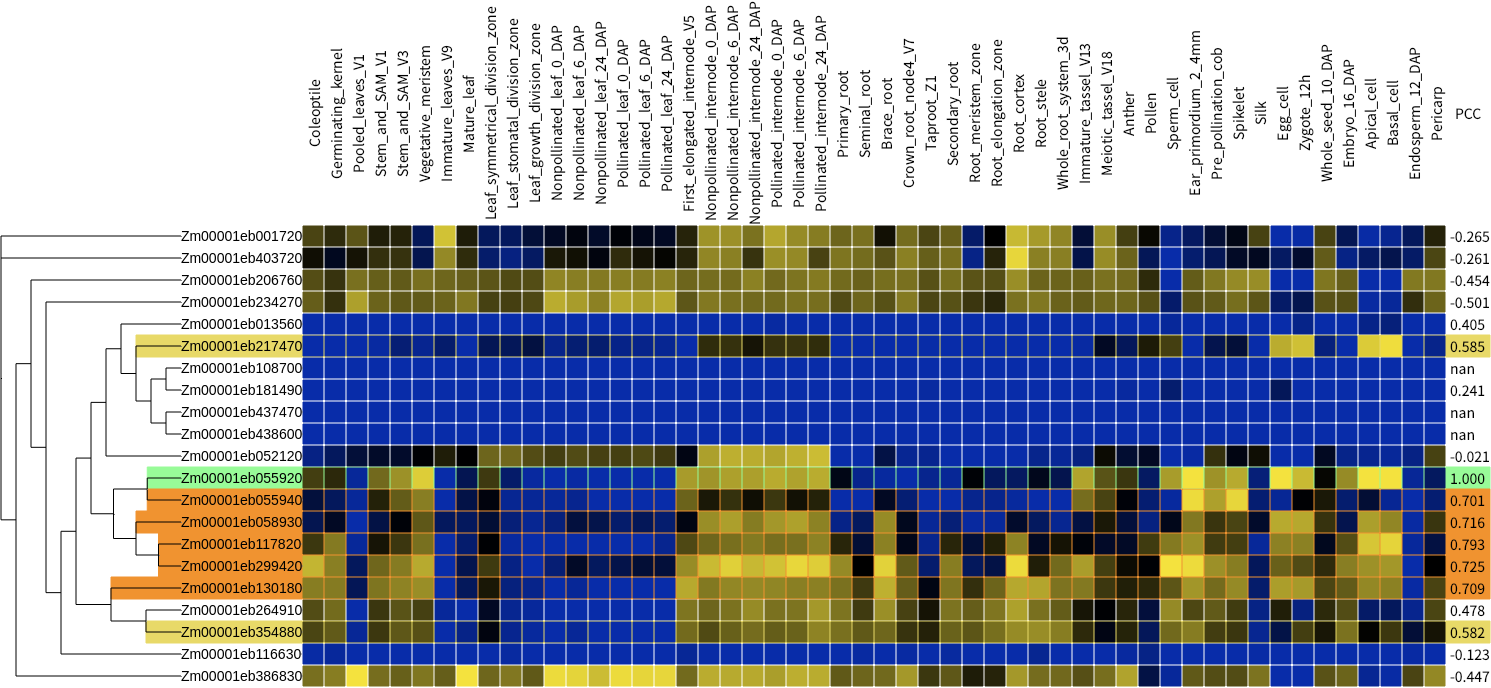
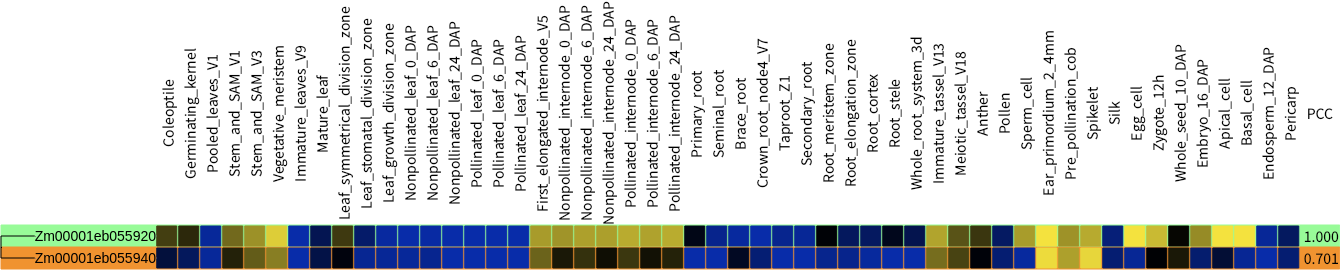
- Color scale log2 expression value

- Pearson correlation coefficient
~ 0.5
0.5 ~ 0.7
0.7 ~ 0.85
0.85 ~
In the era of genome editing, one of urgently required technology in plant biology is finding out proper target which doesn't have functional redundancy. CAFRI is a simple web-based tool that help user to find proper target in genome editing with CRISPR system. It visualizes functional redundancy of quried gene as phylogenetic heatmap and pearson correlation coefficient score. Possible redundant genes are highlighted with yellow, orange, and orange-red background.


| No family | Zm00001eb224330 |
| One family | Zm00001eb077940 |
| Two family | Zm00001eb349990 |
You can get started with CAFRI with the example gene IDs provided above.